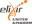General GO annotation
There are two parts to a GO annotation: first, the association between a gene product and GO term; and second, the source and evidence used to make the association.
Each GO term represents an activity, process, or location in the cell of a gene product, and has a name (called the GO term name), a numerical identifier (called the GO ID), and a text definition. The first part of the annotation involves selecting GO term (see choosing the correct GO term).
Choosing the correct GO term (general)
When using GO terms -- or terms from any ontology -- always pay careful attention to the term definitions. They are usually more detailed, and often more informative, than the term names alone. For each annotation, ensure that the definition of the selected term accurately describes the experiment you are trying to capture, and that the results shown in the paper fit all parts of the term definition.
To find a GO term, type text into the search box. When suggestions from the autocomplete feature appear, choose one and proceed. If your initial search does not find any suitable terms, try again with a broader term (some examples are provided in the specific sections below). Selecting a term takes you to a page where you can read the definition to confirm that it is applicable. More specific "child" terms will be shown (where available), and you can select one of these more specific terms in an iterative process. GO terms are organised in a hierarchical structure, and GO annotations should be as specific as possible to describe the data from your experiment. (More information on GO organisation is available on the GO web site.)
See the "Specific branches of GO" sections below for more advice on choosing terms from each ontology aspect (cellular component, biological process, molecular function).
You will have the opportunity to request a new term if the most specific term available does not describe your gene product adequately.
Choosing the Evidence code
After selecting a GO term, you must choose the evidence code that best describes your experiment.
- Mutation(s) in a single gene: IMP (inferred from mutant phenotype)
- Mutations in more than one gene in the same strain: IGI (inferred from genetic interaction)
- Direct physical interaction of one single gene product with another: IPI (inferred from physical interaction)
- Direct assay of the location, complex, function or process: IDA (inferred from direct assay)
Data supporting the evidence
"With" field: IGI or IPI evidence requires that you indicate the interacting gene product. Choose the appropriate gene from the list you initially entered (or add genes to the list if necessary).
Editing, deleting and duplicating GO annotations
Edit: If you want to make changes to an annotation you have made, use the "Edit" link next to the annotation in the table. In the pop-up edit the appropriate fields, then click "OK".
Delete: The "Delete" link will ask you to confirm that you want to remove an annotation, and then delete it.
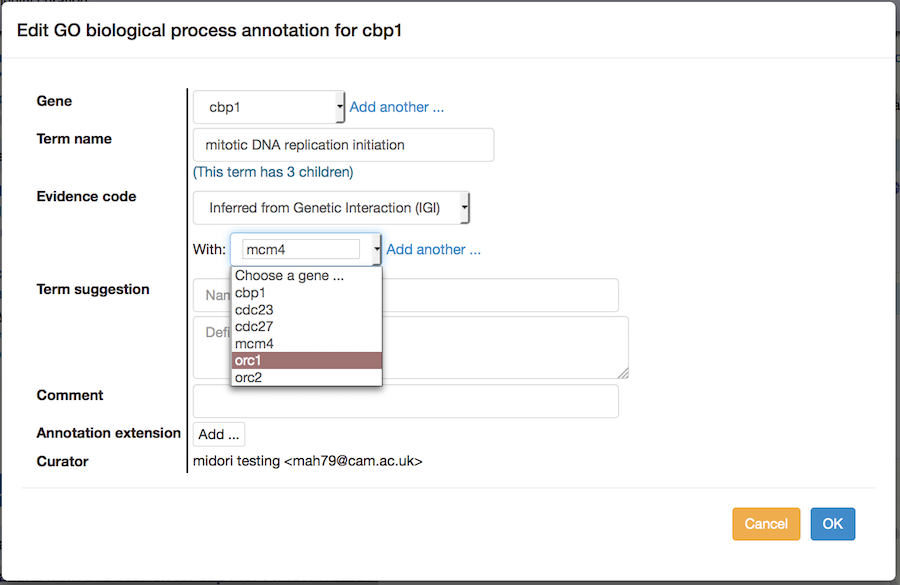
Copy and edit: The "Copy and edit" link in the table on a gene page allows you to make another annotation to the same gene. For example, you may want to indicate that genetic interactions with two different genes both support annotation to the same GO term. The interface works the same way as for editing an annotation, except that a new annotation is created, and the old annotation is retained without changes.
On the paper summary page, the "Copy and edit" link adds one more feature: the annotation can be transferred, with or without other changes, to any other gene in the gene list, by changing the top pulldown:
The "quick add" links available in advanced mode open the editing pop-up without any data entered or selected.
Annotation extensions
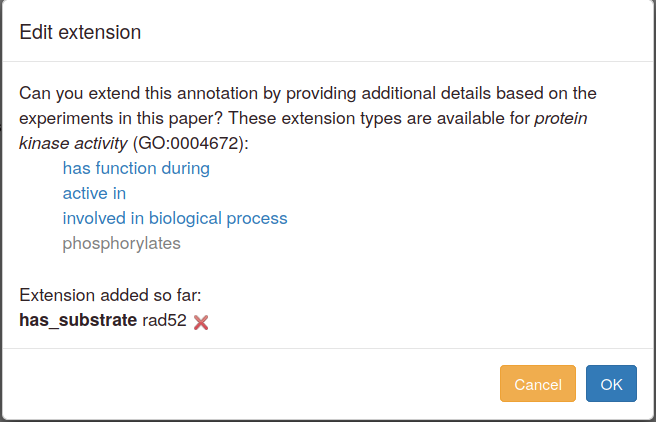
You can add annotation extensions to provide additional specificity for GO annotations. (See the PomBase documentation (and linked GO documentation) for more information on annotation extensions.) After you have selected an ontology term and evidence, the Canto interface will display any available extension types. Click the link to choose an extension type and bring up a pop-up in which you specify the required details for the extension. For example, an annotation to "protein kinase activity" can have any of these extensions:
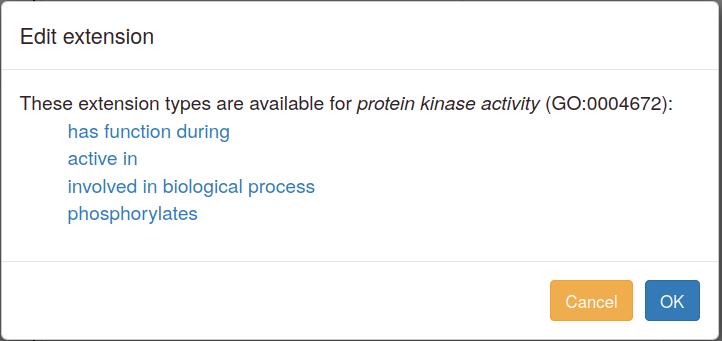
More details on annotation extension options are available in the sections below on specific branches of GO.
If more than one type is offered, you can add one of each type, but if you add them at the same time they will be interpreted as going together to form a compound annotation in which all of the parts apply at once. To create independent annotations (i.e. where one or another may apply, but not necessarily all at once), finish the annotation with one extension (or set of extensions), and then use the "Copy and edit" feature to create another annotation where you can edit the extension(s).
In all cases, the actual relation name used by the database will appear when you have finished the annotation plus extensions.
When you edit or duplicate an annotation, extensions can also be added, amended or deleted. An "Edit" button in the pop-up launches the annotation extension addition steps.

To change an existing extension, first delete it and then add a new one. Editing interface ("protein mislocalized to nucleus" example from FYPO):
Specific branches of GO
Cellular Component
Cellular component describes locations, at the levels of subcellular structures and macromolecular complexes. Examples of cellular components include nucleus, nuclear inner membrane, nuclear pore, and proteasome complex. Generally, a gene product is located in or is a subcomponent of a particular cellular component. The cellular component ontology includes multi-subunit enzymes and other protein complexes, but not individual proteins or nucleic acids.
- Suggested broad terms to start searching
- rough endoplasmic reticulum
- nucleus
- ribosome
- Be careful when interpreting subcellular locations, as certain tagged proteins may be mis-targeted. For example, proteins are often mislocalized to vacuole or other components upon addition of the tag.
- When a macromolecular complex is characterized, all of the subunits should be annotated to an appropriate complex term in the Cellular Component ontology (example, GO:0005681, 'spliceosomal complex' or GO:0000786, 'nucleosome').
Annotation extensions
Any GO cellular component annotation can have extensions indicating when (i.e. in which cell cycle phase or during which biological process, usually a response to stress or another stimulus) a gene product is observed in the given location. For these, type text into the ontology-search autocomplete box, choose from the suggestions, and drill down to more specific terms as usual.
Annotations to "chromatin" or "chromosomal region" terms may also have extensions to specify which regions of a chromosome a gene product is found at. First, use the radio buttons to select a region descriptor (from the Sequence Ontology (SO)) or a gene. For descriptions, type text into the ontology-search autocomplete box, choose from the suggestions, and drill down to more specific terms as usual. Or, choose a gene from the pulldown menu (you can enter new genes at this point if necessary).
Molecular Function
A molecular function is an activity, such as a catalytic or binding activity, that occurs at the molecular level. GO molecular function terms represent activities (protein serine/threonine kinase activity, pyruvate carboxylase activity), rather than the entities (gene products or complexes) that perform the actions. As a general rule, molecular functions correspond to single step activities performed by individual gene products.
- Suggested broad terms to start searching
- transporter
- transferase activity
It is sometimes difficult to distinguish between a Molecular Function and Biological Process term. The key question to ask to establish if you need a Molecular Function term is whether the results shows how the gene product accomplishes its role. For example, if the result shows that a mutant version of a gene product affects transcription, by itself that doesn’t imply that the gene product is a transcription factor. If the study shows that the gene product binds to DNA or protein and thereby modulates transcription, then an appropriate Molecular Function term ('sequence-specific DNA binding RNA polymerase II transcription factor activity' or 'protein binding transcription factor activity') can be used. Data from the mutation experiment can be used to make an annotation to the Biological Process term ‘transcription, DNA dependent’ or to one of its child terms.
Some annotations require careful consideration to ensure that what the experiment shown truly matches the GO term definition. For example, if a paper states in the introduction that a gene product is a transcription factor but provides results only showing DNA binding, this paper should not be used to annotate to 'sequence-specific DNA binding RNA polymerase II transcription factor activity'. The appropriate term would be 'sequence specific DNA binding' or one of its child terms. In another situation, if the author states that a protein is a serine/threonine/tyrosine kinase, but only shows experimental evidence for serine and threonine, the curator can only annotate to 'protein serine/threonine kinase activity'.
Annotating to 'protein binding' (GO:0005515) has to be done with caution as most proteins within the cell bind to other proteins at one time or another. Keep in mind whether the gene product being annotated causes a biologically relevant effect by binding to another protein: if so, protein binding is its function. Only annotate direct protein binding using the GO term "protein binding".
Annotation extensions
Annotations to terms representing catalytic activities that act on protein substrates (e.g. protein kinases or protein transporters) can have extensions that indicate which substrate(s) were used in your experiments. Choose a gene from the pulldown menu (you can enter new genes at this point if necessary).
Use "has function during" to specify a cell cycle phase or another process (usually a response to a stress or another stimulus) in which the activity is observed. When you select it, the ontology-search autocomplete box appears. Type text, choose from the suggestions, and drill down to more specific terms as usual.
For DNA binding terms, "binds region" indicates where the annotated gene product binds. The ontology-search autocomplete box appears, and searches the Sequence Ontology (SO). Type text, choose from the suggestions, and drill down to more specific terms as usual.
Biological Process
A biological process is series of events accomplished by one or more ordered assemblies of molecular functions. It can be difficult to distinguish between a biological process and a molecular function, but the general rule is that a process must have more than one distinct step (e.g. cell cycle, transport, signal transduction).
- Suggested broad terms to start searching
- cell cycle
- transport
- terms in the Biological Process GO slim may be useful starting points
Check that the selected GO term definition matches the specificity of the experiment. For example, if a paper shows experimental results that a gene product can transport specific amino acids and the authors extrapolate that the gene product can transport any amino acid, the gene product should be annotated only to transport of the amino acid that was shown experimentally.
Direct vs. indirect effects: With mutant phenotypes, often it is hard to discern if a gene product is directly involved in a process or if its absence has an indirect/downstream effect. For example if any of the proteins involved in splicing is mutated, it affects translation. This is a downstream effect because most of the genes encoding ribosomal proteins have introns and if splicing genes are mutated, these ribosomal genes are not processed, thereby affecting ribosomal assembly and hence translation. In this case the genes involved in splicing shouldn’t be annotated to translation. Instead, you should make a phenotype annotation to "abnormal translation" or one of its children.
Annotating to "response to stimulus" terms: There are many RNA expression studies that measure the levels of RNA species when exposed to various stimuli and then suggest that the genes are over expressed and thus involved in responding to that stimulus. An increase in expression during a process does not always imply that the genes are directly involved in that process. The ‘response to’ terms are intended to annotate gene products that are required for the response to occur (e.g. production of a gene product or hormone, or initiation of cell division). If nothing else is known about the gene product, make a phenotype annotation to "increased RNA level" or one of its children
Annotating to regulation terms: Regulation of a process is defined as any pathway that modulates the frequency, rate or extent of that process. To decide if the gene product participates directly in a process or regulates the process, consider: does the gene product being annotated perform within the pathway or upstream of the pathway to start or stop or change the rate of the process? Note that a gene product may be involved *both* directly in a process, and in its regulation (for example enzymes in a pathway which regulate the pathway via a feedback loop).
Annotation extensions
Extensions allowed for GO biological process terms fall into a few broad categories:
- Genes/gene products, such as proteins affected by localization or modification processes, or targets of regulatory processes: Choose a gene from the pulldown menu (you can enter new genes at this point if necessary).
- Locations where a process takes place. Most are regions where DNA- or chromatin-based processes occur, and use Sequence Ontology (SO) terms, although a few are locations within the cell, using GO cellular component terms. For these, type text into the ontology-search autocomplete box, choose from the suggestions, and drill down to more specific terms as usual.
- A cell cycle phase or other process (usually a response to stress or another stimulus) during which the annotated gene product participates in the process. For these, type text into the ontology-search autocomplete box, choose from the suggestions, and drill down to more specific terms as usual.