A protein modification is any covalent modification or other change that alters the measured molecular mass of a peptide or protein amino acid residue.
Canto uses the PSI-MOD ontology for protein modification annotations.
When you choose "Protein modification", you go to an ontology term search page that works the same way as those for GO and phenotype terms: start typing a term, and matching suggestions will appear.
You may find it helpful to search for a broad term (e.g. phosphorylated, methylated, lysine), especially if you have trouble finding a specific term.
After you choose a term from the autocomplete list, you will go to a page that shows the term definition and any more specific terms in the ontology. You can thus refine your search iteratively before making your final selection (examples of more specific terms are: O-phospho-L-serine, monomethylated L-lysine). Also see the hints below, which should help you search for, and recognize, the terms you need. Please read the definition carefully to ensure that it fits your results accurately and completely.
When you have decided which term to use, click "Proceed" on the term page. On the next page, select the applicable evidence from the pulldown, then "Proceed" again. On the final page, you can add a comment; for example, if you have any details that don't fit the available annotation extensions, you can put them in the "message to curators" box (which you see when you submit your curation session).
Gene products may be annotated with multiple protein modification terms.
Hints for finding useful modification terms
Because PSI-MOD contains a number of very specific terms that may not be familiar to all Canto users, PomBase curators have assembled a list of common modifications and the best PSI-MOD terms to use for them.
- Phosphorylation: A search for "phosphorylated residue"
will find the broadest term (MOD:00696). You can use this term directly, but
the vast majority of phosphorylation fits the definition of "O-phosphorylated
residue" (MOD:01455). If you know which residue(s) is/are phosphorylated, you
can use one or more of the more specific terms. The most commonly applicable
are:
- phosphorylated serine: O-phospho-L-serine (MOD:00046)
- phosphorylated threonine: O-phospho-L-threonine (MOD:00047)
- phosphorylated tyrosine: O4'-phospho-L-tyrosine (MOD:00048)
- Methylation: PSI-MOD contains many terms decribing methylation, for which the iterative search is particularly helpful. We therefore suggest that you start by searching for the most general term, "methylated residue" (MOD:00427). You can then work your way down to terms of intermediate specificity, such as "N-methylated residue" (MOD:00602), "methylated arginine" (MOD:00658), "methylated lysine" (MOD:00663), etc., and, if applicable, very detailed terms such as "N6,N6,N6-trimethyl-L-lysine" (MOD:00083).
- Glycosylation: The broadest term is "glycosylated residue" (MOD:00693). Although many very specific terms are available in PSI-MOD, we anticipate that "glycosylated residue" or one of the child terms shown on the MOD:00693 page (e.g. "N-glycosylated residue" (MOD:00006), "O-glycosylated residue" (MOD:00396)) will suffice for most protein modification annotation in Canto.
Annotation extensions
You can add annotation extensions to provide additional specificity for protein modification annotations. (See the PomBase documentation for more information on annotation extensions.) After you have selected an ontology term and evidence, the Canto interface will display any available extension types. Click the link to choose an extension type and bring up a pop-up in which you specify the required details for the extension. For example, an annotation to "O-phospho-L-threonine" can have any of these extensions:
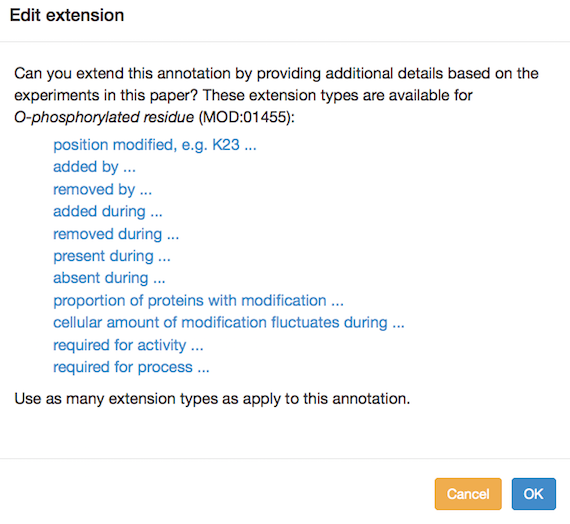
Several annotation extension types are allowed for protein modifications, falling into a few categories:
- for "added by" or "removed by", choose a gene from the pulldown menu (you can enter new genes at this point if necessary)
- for "proportion of proteins with modification", enter a percent value (e.g. 55%) in the text box
- for "position(s) modified", enter one or more residue
designations, using the single-letter amino acid code and a number
for the position (e.g. S77)
- Note: for histones, residue numbering assumes that the initiator methionine is removed. For example, histone H3 (hht1, hht2, hht3) is methylated on residues numbered K4 and K9, not K5 and K10.
- for all other extension types, the ontology-search autocomplete box appears. Type text, choose from the suggestions, and drill down to more specific terms as usual
In all cases, the actual relation name used by the database will appear when you have finished the annotation plus extensions.
You can add one of each type, but if you add them at the same time they will be interpreted as going together to form a compound annotation in which all of the parts apply at once. To create independent annotations (i.e. where one or another may apply, but not necessarily all at once), finish the annotation with one extension (or set of extensions), and then use the "Copy and edit" feature to create another annotation where you can edit the extension(s).
When you edit or duplicate an annotation, extensions can also be added, amended or deleted. An "Edit" button in the pop-up launches the annotation extension addition steps.

To change an existing extension, first delete it and then add a new one. Editing interface:
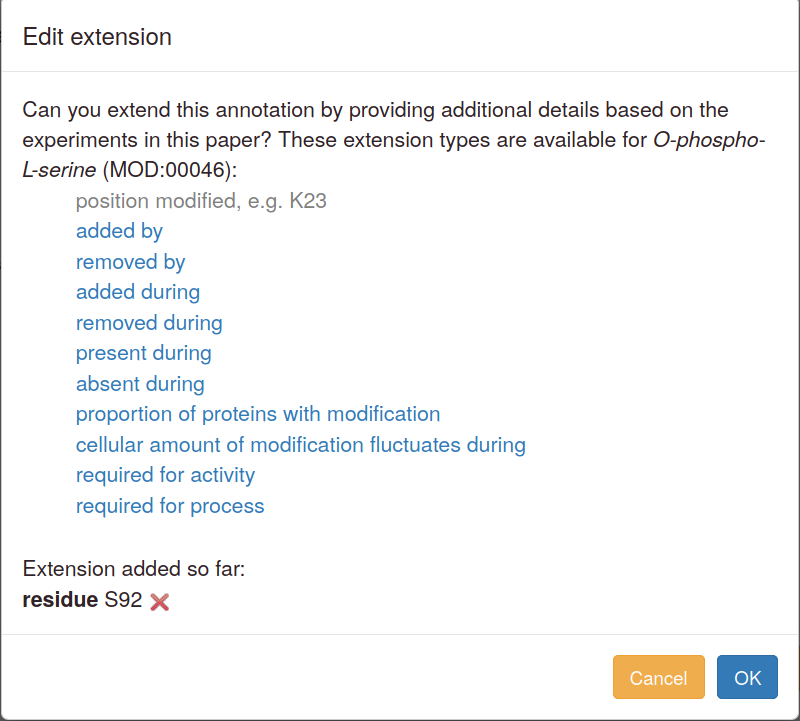
The "quick add" links available in advanced mode open the editing pop-up without any data entered or selected.